Section: New Results
Grouped Local Automated Cell Extractor (GLACE )
Participants : Gaël Michelin, Grégoire Malandain.
This work is made in collaboration with Julien Laussu, Patrick Lemaire (CRBM, CNRS, Montpellier 1 & 2 university), Emmanuel Faure (IRIT, CNRS, Toulouse) and Christophe Godin (Inria Virtual Plants team, Montpellier).
In developmental biology, the embryogenesis study relies in particular on image-based studies. Today, fluorescent confocal microscopy is a means for in vivo imaging of developing organisms at cell level with a high spatio-temporal resolution. To handle such 3D+ image sequences, adapted computer-assisted methods are highly desirable in order to extract essential information from these data.
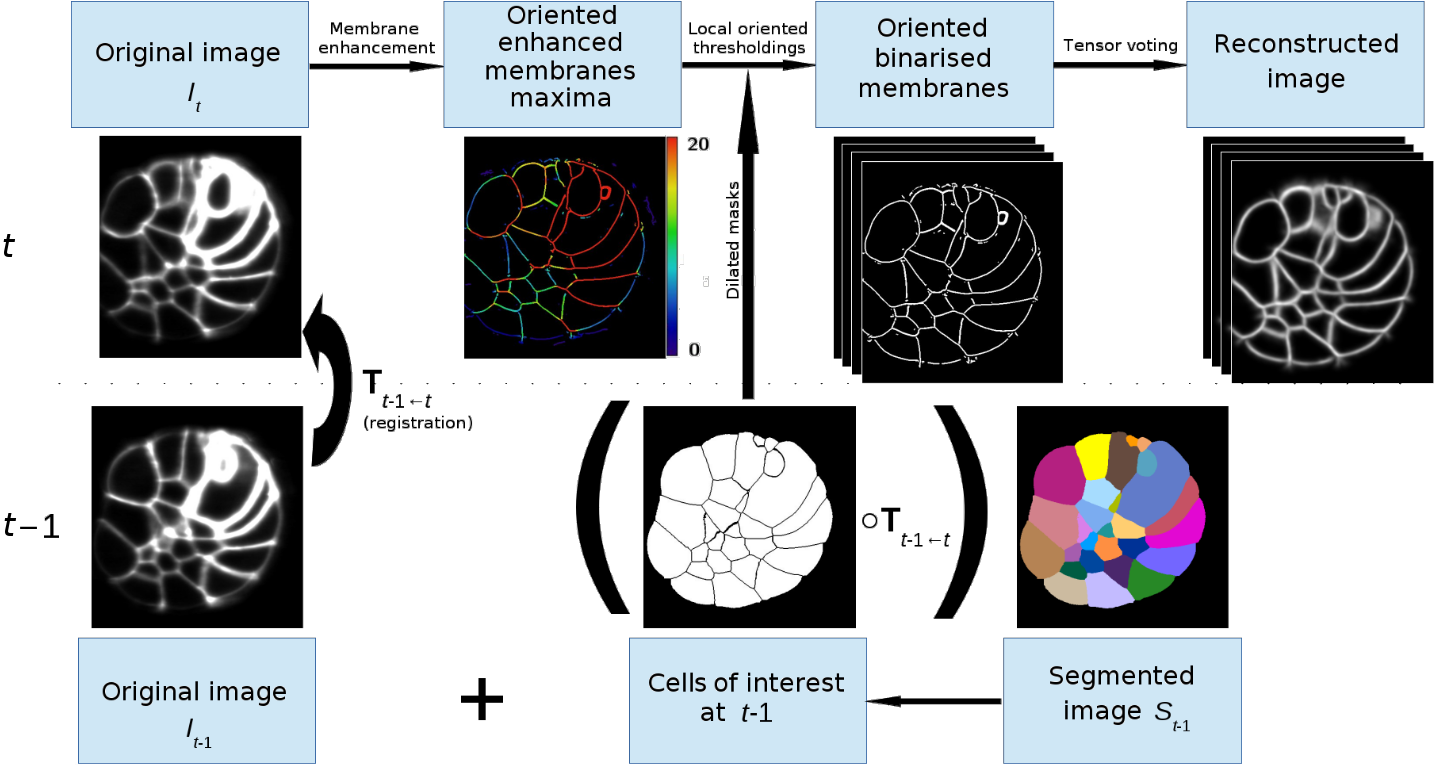
More specifically, for developing ascidian embryos, an existing framework called ASTEC [19] is used by biologists in order to extract the cell segmentation and lineage from some 3D+ sequences. However, remaining issues about segmentation accuracy motivated us to propose a new framework as an alternative to ASTEC for cell segmentation and tracking. The originality of the proposed Grouped Local Automated Cell Extractor (GLACE ) framework is to segment the –th image of a sequence by applying locally the original 3D cell segmentation framework of [21] for all the regions of interest defined by the segmented cells of the –th image of the sequence. The union of all the local reconstructions provides the segmentation of the –th image of the sequence (figure 11). The GLACE framework does not replace the ASTEC framework, however they provide complementary results for embryo image sequence reconstructions.